This is an adaptation of a second workshop I ran in 2022 as part of the Centre of Excellence for the Dynamics of Language (ANU) Seminar Series - I’ve subbed out the data I used in the workshop with the iris data again to make this content more accessible. If you would like a recording of the workshop, which uses vowel data, you can email me at elena.sheard@anu.edu.au
#install tidyverse if you haven't already
install.packages("tidyverse")
#load the tidyverse
library(tidyverse) Workshop Structure
- Introduction to dplyr
- Getting familiar with your data
- Cleaning your data
- Subsetting your data
- Transforming your data
- Summarising your data
- Integrating dplyr and ggplot
Introduction to dplyr
What is dplyr?
- A grammar of data manipulation
- provides a consistent set of verbs that help you solve the most common data manipulation challenges
- Its functions expect tidy data
- Each variable in its own column
- Each observation in its own row

Why should I learn it?
- Good for manipulating data
- Easier than manual changes and in most instances easier than base R
- Highly transferable skill (general data analysis)
- Commonly used by R users = plenty of resources
- Allows you to create a workflow (e.g., create new spreadsheets from old ones without changing the original)
- Integrates with other tidyverse packages (like ggplot)
The %>% operator
- Dplyr can do multiple things to data in a single piece of code
- To do this, we use the
%>%operator- aka the ‘pipe’ operator
- Shortcut is Ctrl/Cmd + Shift + M
- The pipe allows you to do multiple manipulations to the same data (like the + in ggplot2)
- Do this, and then do this - order matters!
Getting to know your data
loading the data
- Run
df <- read_csv("my_dataframe_name.csv")to read in your data files (although I will be using the Iris dataset here) read_csv()is similar toread.csv()- Faster for large .csv files
- Loads data as a Tibble (data format used by dplyr), read.csv() loads data as a regular data frame
- If you ever need to convert a Tibble back to a conventional dataframe you can use
as.data.frame()
glimpse()
glimpse()shows you every column in a data frame, the type of column it is in <>, and the values in of the first few rows of each column<dbl>stands for ‘double’, which is term sometimes used in programming languages for non-integer numbers (when manually changing factors in R you are likely using ‘numeric’ instead e.g., as.numeric(dataframe$column))
spec()also does the same thing, but doesn’t include the values of the first few rows- You can use this to find column names in the activites throughout the workshop
iris %>%
glimpse()## Rows: 150
## Columns: 5
## $ Sepal.Length <dbl> 5.1, 4.9, 4.7, 4.6, 5.0, 5.4, 4.6, 5.0, 4.4, 4.9, 5.4, 4.…
## $ Sepal.Width <dbl> 3.5, 3.0, 3.2, 3.1, 3.6, 3.9, 3.4, 3.4, 2.9, 3.1, 3.7, 3.…
## $ Petal.Length <dbl> 1.4, 1.4, 1.3, 1.5, 1.4, 1.7, 1.4, 1.5, 1.4, 1.5, 1.5, 1.…
## $ Petal.Width <dbl> 0.2, 0.2, 0.2, 0.2, 0.2, 0.4, 0.3, 0.2, 0.2, 0.1, 0.2, 0.…
## $ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setosa, s…distinct()
- Used to find unique elements of a given attribute (i.e., the different levels of a factor or categorical variable)
data %>% distinct(column_a)
iris %>%
distinct(Species)## Species
## 1 setosa
## 2 versicolor
## 3 virginicaCleaning your data
rename()
- Used for renaming columns
- You must specify the new name first
data %>% rename(new_col_name_1 = old_col_name_1)- A useful function if your column names have spaces
- R does not like spaces, and will insert full stops when you load your data
iris %>%
rename(sepal_length = Sepal.Length,
sepal_width = Sepal.Width,
petal_length = Petal.Length,
petal_width = Petal.Width)arrange()
- Sorts your data by individual columns
data %>% arrange(column_a)- Default is in ascending/alphabetical order. To arrange in descending order you can:
data %>% arrange(desc(column_a))
#Sort rows by duration in ascending order
iris %>%
arrange(sepal_length)
#Sort rows by duration in descending order
iris %>%
arrange(desc(sepal_length)) Subsetting your data
select()
- When you want to select specific columns from your data
- There are two approaches:
- Naming the columns you do want
- Naming the columns you don’t want
- To select the columns you do want:
data %>% select(column_a, column_b, column_c)- compared to
data[,c("column_a", "column_b", "column_c")]in base R
- To remove the columns you don’t want:
data %>% select(-column_d, -column_e, -column_f)- I do not know how to do this in base R and did not think figuring it out was worth my time
- You do not need to have the column names in ” ”
- You can also use
:to tell R you want to select all the columns between column a and column xdata %>% select(column_a:column_z)
- If you have a lot of columns with similar names, you can also select them using
contains()data %>% select(contains(match))
- There are other ways to make selection more specific:
ends_with(match),starts_with(match),match(match),num_range()
#select specific columns from 'iris'
iris %>%
select(petal_length, petal_width)
#select all columns from speaker to vowelSpecificContext
iris %>%
select(sepal_length:petal_length)
#select all columns where the name contains 'F1'
iris %>%
select(contains("sepal"))
#select all columns ending with "e"
iris %>%
select(ends_with("dth"))
#select all columns starting with "a"
iris %>%
select(starts_with("s"))filter()
- When you want to select specific rows from your data
data %>% filter(column_a == "level_1")- Compared to
data[data$column_a == "level_1",]in base R
- Use the
%in%operator when you want to specify multiple levels of the columndata %>% filter(column_a %in% c("level_1", "level_2", "level_3"))- Compared to
data[data$column_a %in% c("level_1", "level_2", "level_3"),]in base R
- You can also exlude columns using
!=(not equal to)data %>% filter(column_a != "level_2")data %>% filter(!column_a %in% c("level_2", "level_3"))
- You can filter by multiple columns and levels!
data %>% filter(column_a == "level_1", column_b == "level_2)
- If you want to create a separate tibble/dataframe to analyse, remember to assign to a new object
new_df <- data %>% filter(column_a == "level_1", column_b == "level_2")
#select the rows that have 'setosa' in the 'setosa' column
iris %>% filter(Species == "setosa")
#select the rows that have a sepal length greater than '5' and have 'setosa' in the 'setosa' column
iris %>% filter(Species == "setosa" & sepal_length > 5)
#select the rows do not have 'setosa' in the 'Species' column
iris %>% filter(Species != "setosa")
#select the rows do not have 'setosa' or 'versicolor' in the 'Species' column
iris %>% filter(!Species %in% c("setosa", "versicolor"))Activity 1
- Rename three columns in the data frame (use
glimpse()to choose)iris %>% rename(=, =, =)
- Select the first 3 columns of the data frame
iris_3 <- iris %>% select()
- Select all columns containing the letter ‘a’
iris_a <- iris %>% select(contains())
- Select all columns except the column ‘Species’
iris_noSp <- iris %>% select()
Transforming your data
unite()
- This will combine two columns into a single column
- Will unite based on the specified order, not their order in dataframe
#create new column and keep the originals
iris %>%
unite(species_petal_length, c("Species", "petal_length"), remove = FALSE)
#create new column and remove the originals
iris <- iris %>%
unite(species_petal_length, c("Species", "petal_length")) separate()
- Turns a single character column into multiple columns
- Default is to split column in question based on any non-alphanumeric value (e.g., a space, hyphen, comma), but this can be specified with
sep = ''
#create new column and remove the originals
iris <- iris %>%
separate(species_petal_length, c("Species", "petal_length"),
sep = "_")
#create new column and keep the originals
iris %>%
separate(species_petal_length, c("Species", "petal_length"), remove = FALSE) mutate()
- Used to create new columns, often based on other columns already in the data
data %>% mutate(new_column = condition/value/expression)
- Can also be used to change existing columns without creating a new one
data %>% mutate(existing_column = condition/value/expression)
#create a new column that is the product of F2-F1
iris <- iris %>%
mutate(length_minus_width = sepal_length - sepal_width)
#Modify an existing column that is the log of F1_lob
iris %>%
mutate(petal_width = log10(petal_width))ifelse()
- Commonly used within the mutate() function
- Lets you create an if-else statement without writing multiple lines of code
- Useful if you only want to alter some levels of a factor, or the value in the column is conditional
mutate(new_column = ifelse(condition, "value_1","otherwise_value_2"))
# Create a new column called 'short_petal'
# If the value in the 'petal_length' column is greater than 3, put "long"
# Otherwise put "short"
iris <- iris %>%
mutate(short_petal = ifelse(petal_length > 3, "long", "short"))
# create a new column called 'width'
# If the value in the 'petal_width' column is less than 0.5 then put "thin"
# Otherwise put the same values as in the 'Species' column
iris %>%
mutate(width = ifelse(petal_width < 0.5, "thin", Species)) case_when()
- An alternative to if_else if you want to change many or all levels of a factor that saves on multiple lines of code
#Create a column called "petals" that classifies rows with a petal length
# greater than 3 as "long", between 3 and 1 as "medium"
# and less than or equal to 1 as "short"
iris <- iris %>%
mutate(petals = case_when(petal_length > 3 ~ "long",
petal_length < 3 & petal_length > 1 ~ "medium",
petal_length <= 1 ~ "short")) fct_recode()
- Used to recode data types (good practice is to recode columns when loading data)
- Main data types are integer, double (other numbers), character, factor
- You may need to convert some columns to factors from characters, or vice versa
# Convert some columns to factors
# Across() instructs R to do the manipulation across multiple columns
# Recode species to be capitalised
iris %>%
mutate(across(c(Species), factor)) %>%
mutate(Species_recode = fct_recode(Species, "Setosa" = "setosa",
"Versicolor" = "versicolor",
"Virginica" = "virginica")) %>%
select(Species,Species_recode)Activity 2
- Create a new column that combines the species and sepal length columns, and keeps the original columns
iris <- iris %>% unite(new_column,c("column_a","column_b"),remove=)
- Create a new column that contains the product of speal length multiplied by petal length (use * to indicate multiplication)
iris <- iris %>% mutate(new_column = expression)
- Create a new column where, if the sepal length is above 2, it is ‘short’, and it is otherwise ‘long’
iris <- iris %>% mutate(new_column = ifelse(condition,"value_1","value_2"))
- Create a new column where, if the species is seposa it is changed to ‘Seposa’, and it is otherwise ‘not seposa’
iris <- iris %>% mutate(new_column = ifelse(condition,"value_1","value_2"))
- Create a new column where, if Species is seposa it is changed to ‘Seposa’, but otherwise is the same as the Species column
iris <- iris %>% mutate(new_column = ifelse(condition,"value_1",original_column))
Summary Statistics
summarise()
summarize()orsummarise()operators are useful for creating summary statistics on a dataset (e.g., frequency counts, average values)- Used in combination with other functions (e.g., functions that calculate averages)
- If there is no grouping are performed, the result is a single value
data %>% summarize(column_a = mean(column_a))will calculate the average value in column a for the whole dataframe
#calculates average sepal length for the whole dataframe
iris %>%
summarise(mean(sepal_length))
#calculate average, median, maximum and minimum sepal length
#for the whole dataframe again
iris %>%
summarise(mean(sepal_length),
median(sepal_length),
max(sepal_length),
min(sepal_length))
#You can also create new columns names
iris %>%
summarise(Average = mean(sepal_length),
Median = median(sepal_length),
Maximum = max(sepal_length),
Minimum = min(sepal_length))- If you want to calculate this value across different groups, you can combine
summarise()withgroup_by()
group_by()
- Usually used in combination with other functions such as summarise()
- Groups your data by the levels/categories in the columns you select
data %>% group_by(column_a, column_b) %>% summarize(column_y = mean(column_z))
# Groups the data by gender
# Calculates the mean and median F1 value for each group
iris %>%
group_by(Species)%>%
summarise(Average_sepal_length = mean(sepal_length),
Median_sepal_length = median(sepal_length))Useful functions for summaries
- These can all be used within summarise()
- Center
- mean(): calculate average
- median(): calculate median
- Count
- n(): count number of rows
- n_distinct(): count number of unique rows
- Spread
- sd(): calculate standard deviation
- IQR(): calculate the interquartile range
- mad(): calculate the median absolute deviation
- Range
- min(): minimum value
- max(): maximum value
- quantile(): quantile
- Position
- first(): Extract the first element of a vector
- last(): Extract the last element of a vector
- nth(): Extract the nth element of a vector (specify position with number)
- Logical
- any(): Report whether any elements are TRUE
- all(): Report whether all elements are TRUE
- n() will count the number of rows (i.e., token numbers)
# Count the number of rows per Species
# And count the number of unique petal lengths per speaker
iris %>%
mutate(across(c(Species, petals), factor)) %>%
group_by(Species)%>%
summarise(Count = n(),
Unique_petals = n_distinct(petals)) Activity 3: Summary statistics
- Calculate the average petal length (summarise) for each speciies (group by) and sort in descending order. Which group has the longest petal
iris %>% group_by(,, ) %>% summarise( = mean()) %>% arrange(desc())
- Calculate the standard deviation of petal length for each Species
iris %>% filter(age=="") %>% group_by(,) %>% summarise( = sd()) %>% arrange(desc())
- If you have time, try and find your own summary statistics
Combinging multiple data sets: joining
- The following joins add variables from one dataframe (x) to another dataframe (y) based on (specified) matching observations (values in columns) between the two dataframes
- Common situation: joining metadata information to linguistic information from separate spreadsheets
- Metadata has fewer rows (one per speaker) than linguistic data (multiple rows per speaker)
- It is also fine to use
merge()if you’re already comfortable with itmerge()is capable to doing all of these joins, depending on what you specify for the arguments
- All join functions require you to specify the column(s) by which you want to join the two data frames
- These columns must have the same name
- The values in these columns must also match or the join will not work/be incomplete
left_join():
- Returns all rows in x and all columns from x and y
- Rows in x with no match in y will have NA values in the new columns

Source: Statistics Globe
right_join()
- Returns all rows in y and all columns from x and y
- Rows in y with no match in x will have NA values in the new columns

Source: Statistics Globe
inner_join()
- Returns all rows from x only where there are matching values in y
- Shared observations in both dataframes
- I like to use this join because it is easier to spot where rows have gone missing (usually because the ID value does not match)
merge()function by default performs inner join: it returns only the rows in which the left table have matching keys in the right table

Source: Statistics Globe
full_join()
- Returns all rows in x or y
- All rows and columns from both x and y. NA where there are not matching values.

Source: Statistics Globe
semi_join()
- Return all rows from x where there are matching values in y, keeping just columns from x

Source: Statistics Globe
anti_join()
- Includes all observations in x that do not have a match in y
- Keeps just columns from x
- Can be helpful for identifying mismatched values (e.g., It will extract the filenames that are slightly different between dataframe x and dataframe y and therefore cannot be joined or merged)

Source: Statistics Globe
Integrate with ggplot
- One of my favourite aspects of dplyr is that you can subset data directly into ggplot visualisations
- Meaning you don’t have to be constantly creating new subsetted dataframes
Average midpoint
- Here I want to visualise the average petal length and sepal length for each Species
group_by(Species)to group by Speciessummarise(petal_length = mean(petal_length))to calculate the average petal length values for each group- Move into ggplot and switch to
+instead of pipe
iris %>%
mutate(petal_length = as.numeric(petal_length)) %>%
group_by(Species) %>%
summarise(petal_length = mean(petal_length),
sepal_length = mean(sepal_length)) %>%
ggplot(aes(x = petal_length, y = sepal_length, color = Species)) +
geom_point() +
theme_bw()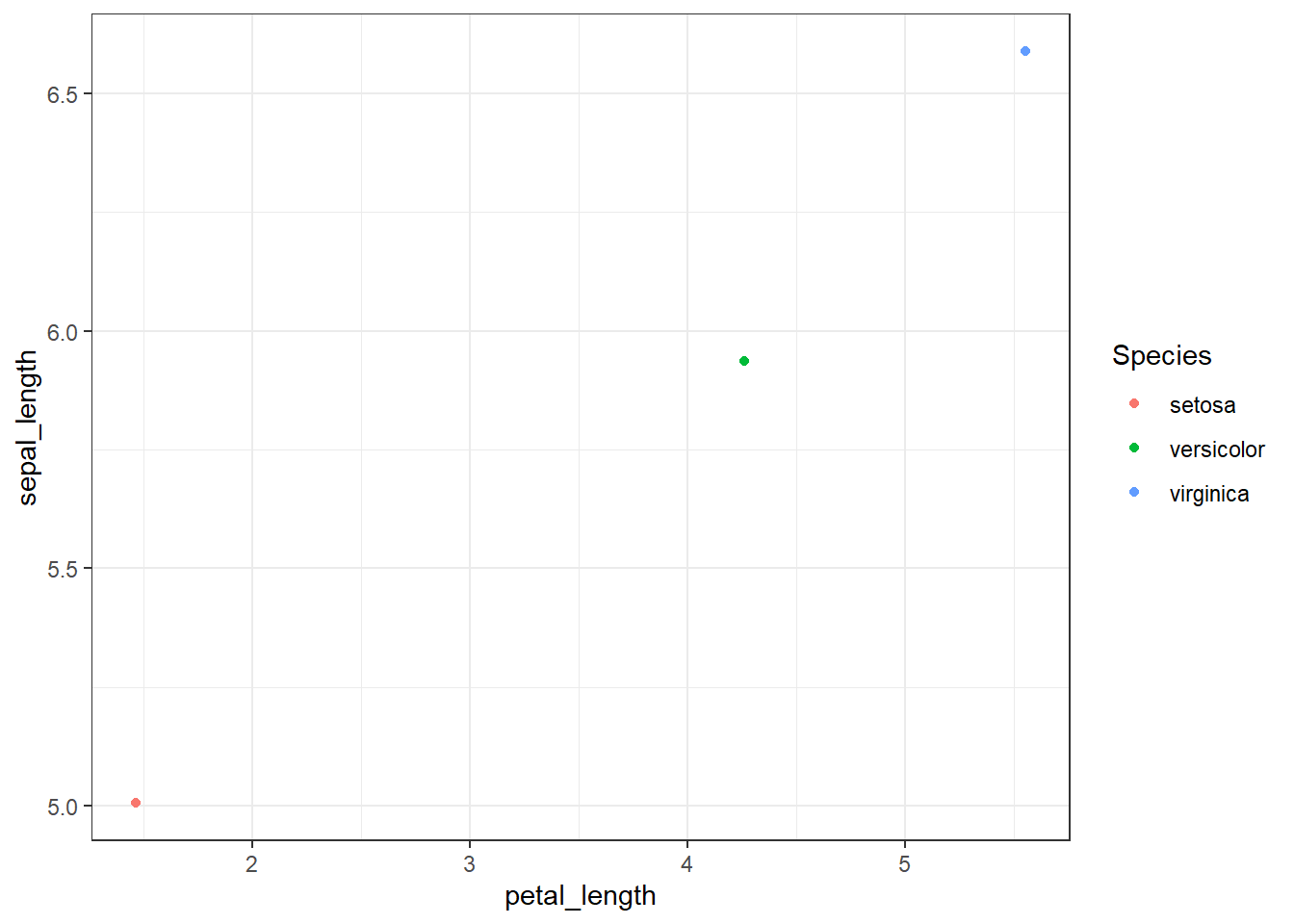
Sepal length
- Boxplot of sepal length for all species except setosa
- Boxplots need all data points (not just averages), so
summarise()is not necessary (but you could filter out a specific point of the vowel)
iris %>%
filter(Species != "setosa") %>%
ggplot(aes(x = Species, y = sepal_length)) +
geom_boxplot() +
theme_bw()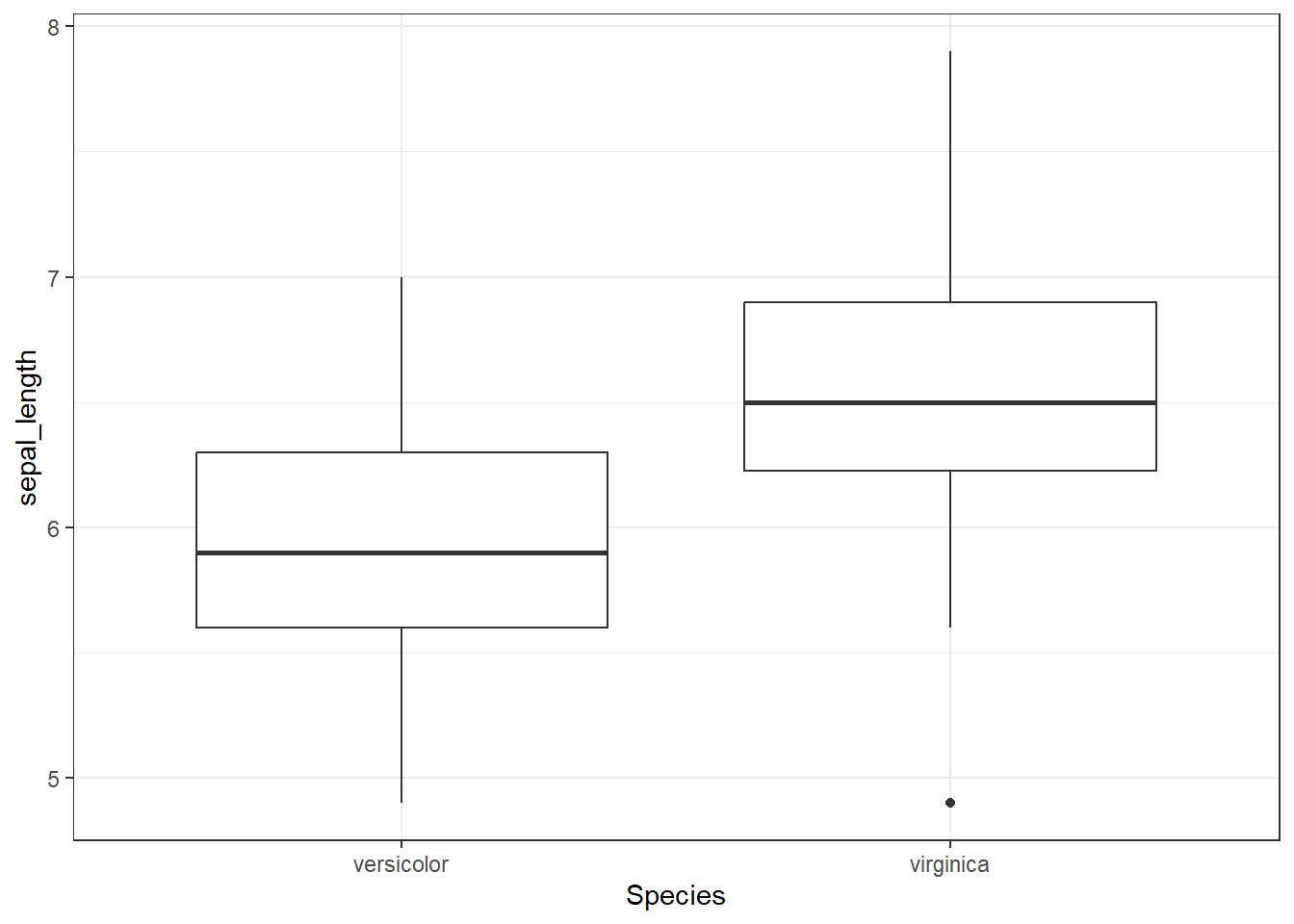
Activity 5
- Try and make your own combination of dplyr functions and ggplot
Sources and resources
https://dplyr.tidyverse.org/
https://r4ds.had.co.nz/transform.html
https://bookdown.org/ansellbr/WEHI_tidyR_course_book/manipulating-data-with-dplyr.html
http://statseducation.com/Introduction-to-R/modules/tidy%20data/gather/
https://rpubs.com/williamsurles/293454
https://www.datasciencemadesimple.com/join-in-r-merge-in-r/#:~:text=We%20can%20merge%20two%20data,database%20join%20operation%20in%20SQL.
https://statisticsglobe.com/r-dplyr-join-inner-left-right-full-semi-anti